Proving I can code enough to study bioengineering - Part 3
The programming challenge:
After learning more about the background of the question, we will move towards the programming part. Luckily I’m not a novice in programming. For this task, I will use Python (duh) and “Atom” as my code editor. I like the Atom interface, and writing on a black background makes me feel more like a professional programmer than I certainly am. A fair warning: I will not share any code, to maintain the integrity of the original challenge.
1) Readout a DNA sequence from a file in FASTA Format
On the NCBI website I searched for bacteria genomes, and opted to use Haemophilus influenzae . The size of this sequence is around 1.85mb, smaller than a lot of others, but also not the smallest. The initial question even asks to use a smaller sequence. It will hopefully help with the runtime of the code.
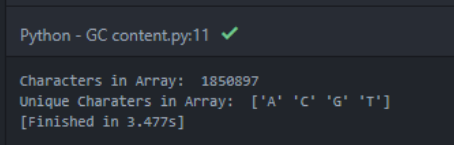
To read the FASTA format I downloaded the Biopython library for Python. My rule of thumb is, that whatever “weird” challenge you think you have at hand, somebody a lot nerdier than you already built a Python library for it. After reading the FASTA file, I plug the DNA sequence into a numpy array to be more easily workable. I then take a first look at the length of the sequence I just downloaded: 1,850,897. Wow, that’s a lot of base pairs. I further confirm the types of unique letter elements we have in the array and find that we are dealing with only “A”, “C”, “G”, “T”.
Here is output of Atom:
I reason that the array of 1.8M ACGT represents one side of the double helix of the genome. As every base can only pair with one specific other base, this connotation is enough. I can deduct the other chain of the double helix. (Having an “A” at a certain spot, means there MUST be a “T” on the other side of the double helix).
Well, this takes care of question 1. YEAH !!
2) Write a function to calculate the G-C content of the genome
We now have to calculate the G-C content as a % of the complete genome. As we know, e.g. with a PCR test the G-C content essentially answers the question: How many “strong bonds” (3 hydrogen bonds; G-C) do we have within a genome compared to how many “weak bonds” (A-T).
As I’m looking into one side of the helix, I know that every “G” Or “C”, is a “strong bond”.. The approach is therefore “simple”:
Count all “G” , Count all “C”, Count all letters. Then calculate: (Sum the count of G + count of C letters) / (Sum of All letters)
Puzzle piece 1: Sum of All letters - A straightforward length function does the trick. We have checked this already: for the specific sample we’re looking at, this is ~1,8M.
Puzzle piece 2: Sum of G and C letters - Here I run two simple numpy count functions. 1x counting all the “G”s + 1x counting all the “C”s.
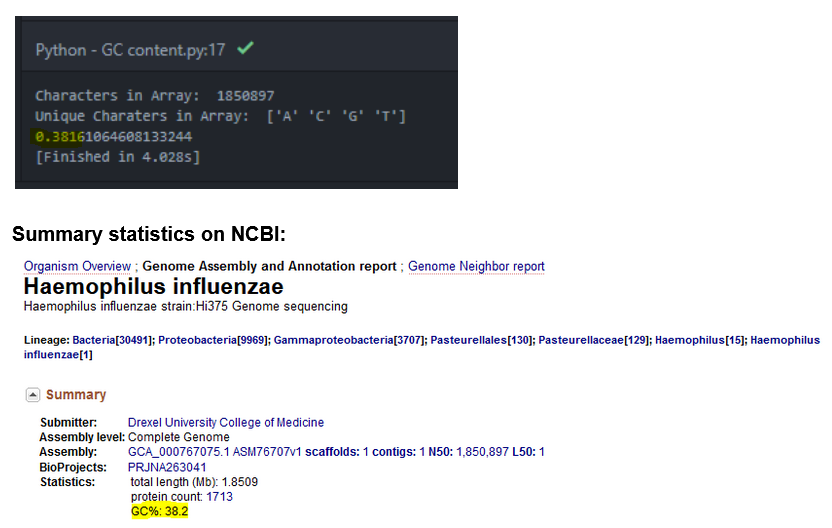
I have the function return count_G+C / len(sequence) to attain my %. Running my function over the influenza genome I receive 0.3816 as an answer. This matches the 38.2% GC content on the summary statistics visible on the NCBI homepage
Question 2 done !!
3) Write a function with a “sliding window” which runs through the genome and calculates the difference of the G-C content of the sample with the average of the genome.
The size of the sliding window should be adjustable through a parameter, the default value is 50
The position with the highest difference should be the output
Now comes the tricky question. First we need to establish what a sliding window is:
It basically takes a cut of the whole sequence according to the size of the window. Then it performs some action with this “subsequence”, and then continues with the next subsequence. In the below graphical example, we have a total sequence length of 10 characters and a sliding window size of 3.
So we take a 3 bite-sized cut of the data, e.g. calculate the difference in G-C content of this subsequence vs. the whole genome. When we’re done calculating we jump to the next subsequence. In our programming challenge, the size should be adjustable with a standard of 50.
Writing it down in Peusdocode I’m thinking of it as follows:
Calculate the G-C content of the whole input sequence (in our case the 1.8M records with a G-C content of 38.2%).
(here we can leverage the function of question 2)
Create a “subsequence” in the size of the sliding window (e.g. 50 letters long) and calculate the G-C content of this subsequence (again, we can use our function from question 2, as the function is sequence length independent)
Calculate the difference between the G-C content of the subsequence with the overall G-C content
If the calculated absolute difference is the largest yet seen, save this difference + the location. Continue the next subsequence
If the calculated difference is not the largest yet seen, do not save. Continue with the next subsequence
Iterate through the whole genome and return the largest seen difference and location at the end
I will refrain from posting any code here, as the university I’m taking this example from would surely not be happy about it. All I can say, that I now have this code available and found it relatively straightforward to code. Of course, I was using Google etc. to find ideas for solutions of certain code segments. For me, there is no shame in that, as we don’t need to re-invent the world here (and have you seen all the memes programmers make about copy & pasting code from stackoverflow?).
I was using the Biopython and numpy libraries to solve this. Using the 50 sized sliding window of the initial question, my answer is:
If anyone gets a different answer, please contact me and we can compare code / find out how mine is wrong !
With that, I’m leaving this be. I captured another snapshot of chaos.